New technology to understand cell types and how diseases develop
An ongoing effort to create detailed molecular atlases of individual cells in different tissues aims to better understand how diseases develop. Now, a team of researchers from Yale and Karolinska Institutet, has developed a technology that brings that goal one step closer. The findings were published in Nature.

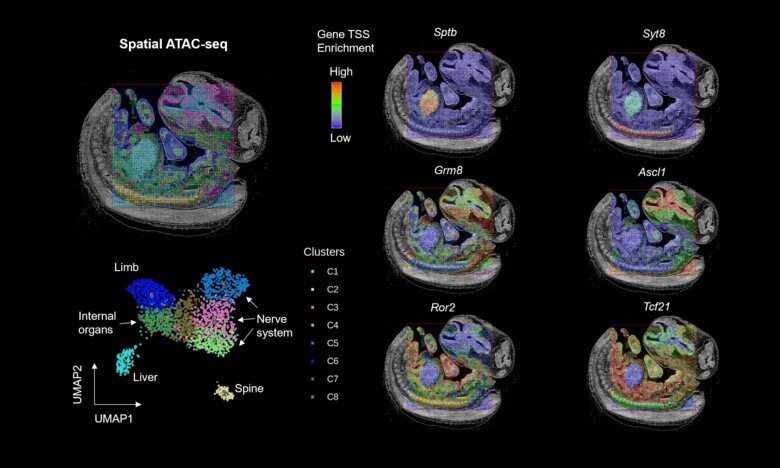
How cells function in tissue depends upon their local environments. Mapping the molecular properties of cells while acquiring their exact location within a tissue is essential for a better understanding of disease. Rong Fan, professor of biomedical engineering at Yale, and Gonçalo Castelo-Branco, professor of glial cell biology at the Department of Medical Biochemistry and Biophysics, Karolinska Institutet, led a team of researchers in developing a new technology to do this.
It allows them to define which regions of the chromatin – the complex of DNA and proteins packed within the nucleus of a cell – are accessible genome-wide in cells at specific locations in a tissue. This chromatin accessibility is required for genes to be activated, which then provides unique insights on the molecular status of any given cell. Combining the ability to analyze chromatin accessibility with the spatial location of cells is a breakthrough that can improve our understanding of cell identity, cell state and the underlying mechanisms that determine the expression of genes – known as epigenetics – in the development of different tissues or diseases.
Building a spatial cell atlas
“Now we can identify the cell types to build a spatial cell atlas based in chromatin accessibility,” Fan says in a news release from Yale University. “We can directly see the cell types at an epigenetic level either for a better definition of cell states or the discovery of cell types.”
The researchers profiled both mouse and human tissues using a technique known as “spatial-ATAC-seq.” Applying this technique to brain tissue revealed the intricate development process of different brain regions. They also applied it to the human tonsil tissue, which provided insight into the organization of immune cell types.
“We’ll get an unbiased global view, and a much finer resolution view, of all possible cell states, and more importantly, ‘see’ where they are in a tissue,” Fan said. “It’s a powerful tool for building cell maps and cell atlases.”
Identifying epigenome cell types
Yanxiang Deng, a postdoctoral associate in Fan’s lab and lead author of the study, said that by using the new method, they were able to identify the epigenome of cell types in the mouse brain tissue in their native location.
“Applying spatial ATAC-Seq in diseased tissues might allow us in the near future to identify transitions between epigenetic states in specific cells in the context of the disease niche, which will give insights of the molecular mechanisms that mediating the acquisition of pathological cellular states,” added Castelo-Branco.
An ambitious global initiative has been undertaken to map out all the different cell types across all of the human organs and different tissue types. Single-cell sequencing has been critical to this effort but it is hard to map the location of cell types to the original tissue environment. This work for the first time allows for directly observing cell types in a tissue as defined by global epigenetic state.
This news article is based on a news release from Yale University.
Publication
Spatial profiling of chromatin accessibility in mouse and human tissues.
Deng Y, Bartosovic M, Ma S, Zhang D, Kukanja P, Xiao Y, Su G, Liu Y, Qin X, Rosoklija GB, Dwork AJ, Mann JJ, Xu ML, Halene S, Craft JE, Leong KW, Boldrini M, Castelo-Branco G, Fan R
Nature 2022 Aug;():
