New method allows analysis of single neurons from tissues
Scientists at Karolinska Institutet in Sweden have developed a new technique that enables them to isolate single neurons from tissues and analyse the gene expression within.

The study, which is published in Nature Communications, demonstrates how the method can be used by neurologists to distinguish between closely related neurons that may show differential vulnerability to disease. The method can also be of value to other areas, such as cancer research, pathology and biomarker identification.
“Similar analyses used to require hundreds, if not thousands of cells,” says Eva Hedlund at the Department of Neuroscience, who led the study with Qiaolin Deng from the Department of Cell and Molecular Biology at Karolinska Institutet. “Our methodological development allows us to effectively analyse a small number of cells, even single neurons, from human and mouse tissues.”

The ability to identify gene expression in different cell populations is a valuable tool for researchers in their efforts to understand disease mechanisms and biological processes. Two hurdles they commonly face is a lack of tissue material, especially from patients with different diseases, and small cell populations. In many cases, there are no unique genetic markers that can be used to identify the cells. Methods that enable identification and analysis of a small number of cells in scarce tissues are therefore warranted.
The teams at KI have now combined two existing methods to develop a new method they call LCM-seq. First they use laser capture microscopy (LCM) to dissect specific cells from tissue samples and then deep RNA sequencing to study all the active genes within these.
“One great advantage of the technique is that the tissue does not need to be broken up,” says Dr Qiaolin. “Thanks to that, the positional information remains intact in each isolated cell. Given that we don’t have to purify RNA molecules from the cells and can use off-the-shelf reagents, LCM-seq is simpler, cheaper and has much higher reproducibility and sensitivity than previous methods.”
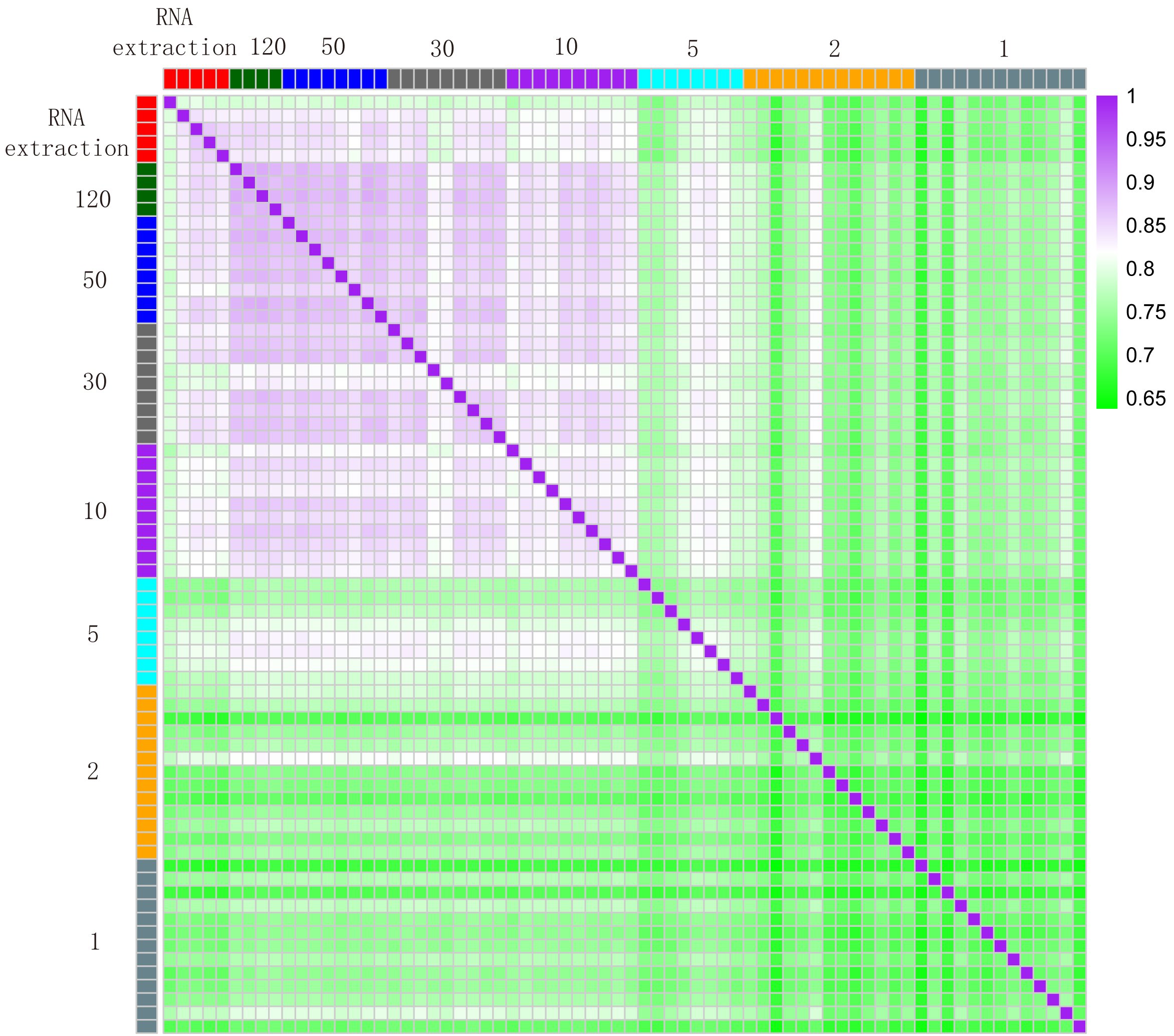
The researchers also show that LCM-seq can be used to learn more about populations of very similar nerve cells. They studied motor neurons isolated from different parts of the spinal cord that are susceptible to degeneration in the incurable disease ALS, and examined dopamine cells from different parts of the midbrain that are destroyed in Parkinson’s disease.
“We show that we can identify specific differences between dopamine cells that are liable to atrophy in Parkinson’s disease and those that are resistant,” says Dr Hedlund. “Since very few cells are needed for LCM-seq, the technique could be of considerable use in other areas, such as cancer biology, pathology and biomarker identification.”
The study was financed by grants from several bodies, including the EU’s Joint Programme for Neurodegenerative Disease, the Ragnar Söderberg Foundation, the Swedish Research Council, Karolinska Institutet, the Åhlén Foundation, the Parkinson Foundation, Neuro Sweden, the Swedish Society for Medical Research, the Jeansson Foundations and the Swedish Foundation for Strategic Research.
Publication
Laser capture microscopy coupled with Smart-seq2 (LCM-seq) for precise spatial transcriptomic profiling
Susanne Nichterwitz, Geng Chen, Julio Aguila Benitez, Marlene Yilmaz, Helena Storvall, Ming Cao, Rickard Sandberg, Qiaolin Deng, Eva Hedlund
Nature Communications, published online July 8, 2016, doi: 10.1038/ncomms12139
